August 12, 2021
An SLAS Technology special collection, “Emerging Trends in 3D Cell Culture: High-Throughput Screening, Disease Modeling and Translational Medicine,” reveals the latest advances in organoid and spheroid research.
Five to 10 years ago, the big hurdle 3D cell cultures needed to clear was convincing researchers of its relevance in life sciences. “Now the idea of studying the human body in its dimensional context has taken hold, and the new challenge is exploring the lab protocol, techniques and peripheral technology that enhance and expand 3D research,” says SLAS Technology Editor-in-Chief Edward Kai-Hua Chow, Ph.D., of the National University of Singapore (NUS, Singapore).
“We live in 3D world – our bodies and organs are not flat like a piece of paper. Why would we study samples that are 2D?” continues Chow, who organized the special collection. “While working in 3D is the best method, it is a different system than the 2D research classically used for the past 40 to 50 years. Life sciences research and discovery must adapt to and develop a whole different set of techniques and technology to capitalize on 3D research.”
To this end, a special collection of SLAS Technology features six articles to help researchers investigate advancements in the 3D space. Covering a range of topics from protocols for controlled cell seeding, the splitting and expansion of human fibroblasts, analyzing neural progenitor cells (NPCs) and customizing induced pluripotent stem cells (iPSCs), the issue focuses on multiple platforms, including microfluidic and high-content imaging formats, that lend themselves well for research on patient-derived cellular models for precision medicine.
“The special collection captures the reader’s imagination and interest as the authors expand what the technology can do,” Chow explains. “These articles offer information for growing complicated 3D organoids, miniaturizing and automating experiments and methods for simply saving time and money by streamlining processes.”
He adds that the future of 3D cell culture research will include multi-cellular organoid cultures and mixing different types of cells together “to show how cells interact as mini organs. Understanding those interactions in an in vitro model can be powerful,” Chow says. “The next step will be to develop assays and technology to the point where any biomedical researcher can take tools off the shelf to create organoids to answer the questions they are trying to ask.”
Chow hopes readers will embrace 3D organoid research. “For those who have already made the jump, there are a lot of resources out there including the content found in SLAS Technology and SLAS Discovery that allow you to make better use of organoids in the lab.”
Capturing Big Ideas in a Miniature Format
An aspect of the special collection that makes 3D cell culture particularly relevant for the SLAS audience, according to Chow, is the use of miniaturization and automation to manipulate organoids for high-throughput screening (HTS). He cites examples from the work of Kenneth Ndyabawe, Ph.D., of the University of Georgia (UGA; Athens, GA, USA), and Anna Popova, Ph.D., at the Karlsruhe Institute of Technology, Institute of Chemical and Biological Systems (Eggenstein-Leopoldshafen, Germany).
“These examples show that 3D research makes lab life easier and more efficient for drug developers and researchers,” says Chow. “Popova shrinks down screenings to use only 100 cells and 30 picomoles of a drug per individual nanoliter-sized droplet. Ndyabawe describes a spheroid chopping technique that allows suspension cultures to be treated like adherent cultures with minimal loss of spheroids due to aspiration.”
In the article, "Spheroid Trapping and Calcium Spike Estimation Techniques Toward Automation of 3D Culture," Ndyabawe and fellow authors from UGA’s School of Chemical, Materials and Biomedical Engineering, College of Engineering, Driftmier Engineering Center and School of Electrical and Computer Engineering, present a device that is compatible with traditional tissue culture plates and confines microtissues in a small area. The team illustrates an automated morphology-independent procedure for cell recognition, segmentation, and a calcium spike detection technique for high-throughput analysis in 3D cultured tissue. Since the recognition and segmentation technique is independent of morphology, the team’s protocol provides a versatile platform for the analysis of large confocal calcium imaging data from several cell types, including neuronal and glial cells.
In "Miniaturized Drug Sensitivity and Resistance Test on Patient-Derived Cells Using Droplet-Microarray," Popova et al. describe miniaturizing experiments using a novel droplet-microarray platform based on a hydrophilic-superhydrophobic patterned surface. The extremely miniaturized platform demonstrates that the dose response of as few as 100 primary patient-derived chronic lymphocytic leukemia (CLL) cells to anticancer compounds resembles the dose–response obtained in 384-well plates requiring 20,000 tumor cells per experiment. The collaboration of authors, representing institutes located in Heidelberg, Germany, including the National Center for Tumor Diseases, the Molecular Medicine Partnership Unit and the European Molecular Biology Laboratories, shares how the platform carries great potential for ex vivo drug sensitivity and resistance tests on patient-derived tumor cells and potentially for implementing such tests for precision medicine.
Chow also mentions that details captured in the special collection make it meaningful to those wanting to delve deeper into 3D techniques, such as the step-by-step protocols found in "Three-Dimensional Macroporous Sponge for the Culture of Hepatocellular Carcinoma (HCC) Patient-Derived Xenograft (PDXs) Organoids," from lead author Tan Boon Toh, Ph.D., who represents both the N.1 Institute for Health and the Institute for Digital Medicine (WisDM) at the Yong Loo Lin School of Medicine at NUS (Singapore).
“Anyone can follow this protocol for developing and using sponge material to improve the culture of organoids,” Chow comments. “This is important because depending on what type of organ you are using to develop the 3D structures, not all of them can grow in liquid. Some of these organoids require a secondary scaffold to grow or some kind of frame in which they can build.”
Tan and fellow authors reveal a method for growing liver cancer organoids through the use of a previously developed microporous sponge scaffold that provides biochemical and mechanical cues to support the culture of normal hepatocytes as spheroids with maintained functionality. Leveraging their success using this sponge scaffold in vitro, the group demonstrates that a similar sponge scaffold enables the maintenance of HCC PDX cells as organoids with preserved viability, molecular features and heterogeneity. The collaborative group represents several institutes at NUS, including the Department of Physiology at WisDM, the Mechanobiology Institute and Department of Biomedical Engineering, the CAMP, Singapore-MIT Alliance for Research and Technology, the Institute of Bioengineering and Nanotechnology and the Agency for Science, Technology and Research (A*STAR).
“For many researchers, developing 3D techniques can be a challenge,” Chow says. “You attempt following the materials and methods section of published papers, but sometimes those sections aren’t always complete, or it’s a trade secret the research team can’t share. If you’re fortunate, you learn techniques from other experts who are willing to share methods.”
This is the case in a research article from Evan F. Cromwell, Ph.D., and fellow authors from Protein Fluidics (Burlingame, CA, USA) and Molecular Devices (San Jose, CA, USA), which describes methods for a novel automated organoid assay system (the Pu·MA System) that is combined with microfluidic-based flowchips to facilitate 3D cell-based assays. "Disease Modeling with 3D Cell-Based Assays Using a Novel Flowchip System and High-Content Imaging” reveals how the assay enables 3D cell-based cultures to mimic in vivo conditions, performs multidosing protocols and multiple media exchanges, provides gentle handling of spheroids and organoids and allows a wide range of assay detection modalities.
“Microfluidics looks at biology in the context of flow-through, because liquids can go in and out of the chip and allow you to do more, such as multiple dosing protocols,” Chow explains. “The idea is that if you have a miniature body system, you can exchange different drugs, different media and see how the organoids respond. Having a clear, optically compatible bottom allows for high-content imaging and detailed analysis of the organoids in these chips. The beautiful images captured by Cromwell’s team show how the technology is able to reveal drug or stress response in the context of not just markers of whether the cell is alive or dead, but also how the structure changes shape and moves in response to those external stimuli – that’s significantly valuable.”
Another article in the special collection, from Pouria Rafsanjani Nejad, M.Sc., and fellow authors from the Department of Biomedical Engineering, The University of Akron (Akron, OH, USA), highlights the idea that 3D organoid research “is not reserved for cancer cells or disease models – it allows for more realistic toxicity testing in normal cells,” Chow says. “How to make drugs safer is just as important as using these systems to study how to better treat diseases.”
Nejad et al. describes in "Toxicity of Combinations of Kinase Pathway Inhibitors to Normal Human Cells in a Three-Dimensional Culture" the toxic effects of combinations of cancer drugs to normal cells in three-dimensional cultures to facilitate more informed treatment selections for subsequent animal studies. The team addresses the challenge of resistance to single-agent chemotherapy and molecularly targeted drugs that prevent sustained efficacy of treatments using a cellular spheroid model to study toxicities of drug combinations to human bone marrow and colon cells. The results indicate that mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase–protein kinase B (PI3K/Akt) inhibitors used simultaneously were only moderately toxic to bone marrow cells but significantly more toxic to colon cells. Molecular analysis of proliferative cell activities and housekeeping proteins further corroborate these results.
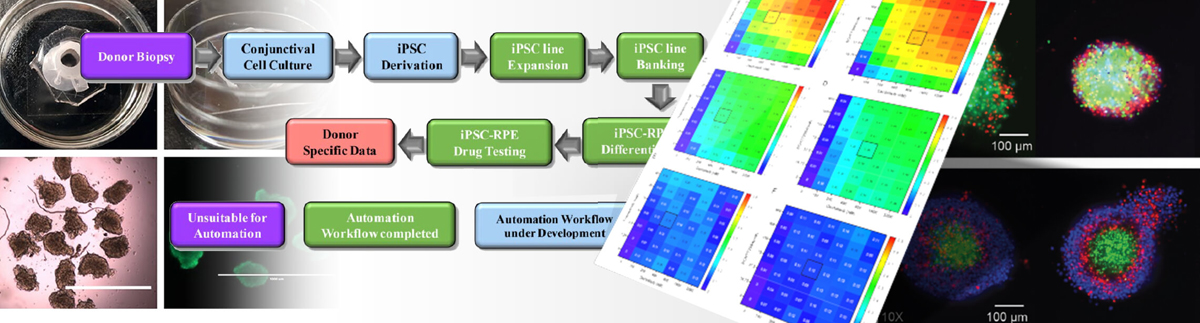
In the article "Automating Human Induced Pluripotent Stem Cell Culture (hiPSC) and Differentiation of iPSC-Derived Retinal Pigment Epithelium for Personalized Drug Testing," research teams demonstrate scalable, reproducible culture and differentiation of hiPSCs to generate medically important cell types from individual patients and patient populations for research and the development of potential cell therapies.
Lead author Vincent Truong, B.Sc., MBA, and fellow authors from the University of Minnesota’s Stem Cell Institute, Department of Ophthalmology and Visual Neurosciences and Department of Genetics, Cell Biology and Development (Minneapolis, MN, USA), use TECAN Fluent automated cell culture workstations to perform hiPSC culture and differentiation to generate patient-derived retinal pigment epithelial cells for downstream use, including drug testing. hiPSCs derived from multiple donors with age-related macular degeneration (AMD) were introduced into the automated workflow, and cell lines were cultured and differentiated into retinal pigment epithelium (RPE). Donor hiPSC-RPE lines were subsequently entered in an automated drug testing workflow to measure mitochondrial function after exposure to “mitoactive” compounds.
Learn More
Explore free online access to the SLAS Technology special collection, “Emerging Trends in 3D Cell Culture: High-Throughput Screening, Disease Modeling and Translational Medicine,” at SLAS Technology online courtesy of the collection’s sponsor, Corning Life Sciences.
